Code Inspection
Overview
Teaching: 0 min
Exercises: 0 minQuestions
What is the purpose of a code inspection?
Which elements of code support reproducibilty?
How do these elements of code support reproducibility?
Objectives
Understand how code documents the analytical workflow
Identify the essential metadata that should be included in a code header to support reproducibility
Explain the importance of non-executable comments for reproduciblity
Understand what a main file is and how it contributes to reproducibility
Evaluate code quality based on whether or not it upholds the reproduciblity standard of being both independently understandable and reusable
Assessing the reproducibility of scientific claims involves an exhaustive list of curation actions, which are outlined in the Data Quality Review (DQR) framework introduced in Lesson 2: Curating for Reproducibility Workflows of the Curating for Reproducibility Curriculum. DQR organizes these curation actions into four components–file review, documentation review, data review, and code review–each of which promote reproducibility by ensuring the completeness, accuracy, understandability, accessibility, and usability of the research compendium.
Code review, in particular, puts reproducibility to the test by verifying whether or not repeating the analytical steps used by the original researcher produces the same results reported in the published study. This assessment of reproducibility begins with a thorough code inspection.
Code Inspection Overview
Ideally, researchers, who know their work best and have direct access to all the necessary details of their analytical workflow, would inspect their code before they share it to be sure that all of the scripts needed to load software packages, import and transform data, and produce outputs are included in code files. They would review their code to be sure that computing environment requirements and the expected runtime are specified in code header metadata and that non-executable comments are included throughout the code to serve as signposts for the analytical workflow by indicating the purpose of commands. They would also confirm that their files render properly, and re-run their code (preferably on a machine other than the one used to perform the original analysis) to be sure that the code compiles properly and executes the analysis from beginning to end without errors.
Unfortunately, it is not always the case that researchers review their code for understandability and executability prior to packaging it into the research compendium and sharing it. Thus, the first step of any reproducibility assessment is to inspect the code and confirm that it includes the following:
- Sufficient information to enable users to re-create the computing environment originally used to run the analysis and produce the reported results
- Details about the analytical workflow that allows others to understand and re-trace the computational steps to produce expected outputs
- Scripts for installing necessary software tools and importing data that take into account independent reuse of the code by researchers other than those who initially wrote the code and ran the analysis
The Analytical Workflow
By its very nature, the analytical workflow of computational research is captured in the code that contains the commands to install software package dependencies, import and transform data, and execute scripts to analyze data and produce results.
It is also the nature of research that analytical workflows are rarely linear, which is often reflected in code that produces outputs in a different order than the order in which they are presented as research results in the manuscript. This makes it necessary for the code to provide signposts to allow others to retrace the analytical steps. These signposts should appear in the code as non-executable comments found in code header metadata and throughout the code.
Code Header Metadata
When inspecting the code, it should be evident the technical requirements for running the code successfully. This information is presented as code header metadata, which is a block of non-executable text positioned at the top of the code before any command scripts.
Below in an example of code header metadata:
************************************
*Title of the paper: Smallholder Farmers and Contract Farming in Developing Countries
*DOI: https://www.pnas.org/doi/full/10.1073/pnas.1909501116
*Authors: Eva-Marie Meemken and Marc F. Bellemare
*Corresponding author: Eva-Marie Meemken
*Contact details: Cornell University and University of Minnesota.
*Warren Hall, Ithaca, NY 14850.
*Phone: +1 607 319 1121. email: emm253@cornell.edu
*Code last updated: 11/01/2019
*Software and version: STATA 15 MP, Run on Windows 10, 64-bit
*Code last executed for maintenance review:
*-22 APR 2022
*-Software and version: STATA 17 MP
*-Operating system: Microsoft Windows Server 2019 Datacenter, Version 10.0.17763 Build 17763, 64-bit
*-Processor: Intel(R) Xeon(R) CPU E5-4669 v3 @ 2.10GHz, 2095 Mhz, 10 Core(s)
*-Processing job completed in under 2 minutes using the above computing environment.
************************************
Code header metadata should provide enough information to allow others to recreate the computing environment used to run the analysis. It should also specify acceptable uses of the code with a license, and provide contact information for the creator(s) of the code in the event that challenges arise when attempting to reproduce the analysis.
Checklist: Code Header Metadata
This checklist outlines the specific information that should be included in code header metadata for every code file. A code inspection should confirm the presence of each item in the checklist.
- Formal citation for the article that presents the results produced from the code
- Author contact information including email, affiliation, and ORCID
- Code license that specifies allowable uses and conditions for use of the code
- Computing environment specifications:
- Operating system and version
- Number of CPUs/cores
- Size of memory
- Statistical software package and version
- Packages, libraries, and other software dependencies and their versions
- File encoding
- Date that the code was last updated
- Date that the code was last run successfully
- Estimated time to run the code from beginning to end
Exercise: Code Header Metadata Inspection
Review the sample code and identify each piece of information outlined in the Code Header Metadata Checklist.
Solution
solution
Non-executable Comments
The code header metadata is an example of non-executable comments that also appear elsewhere in the code. Non-executable code comments are lines of human-readable text that can be placed throughout the code without interfering with command scripts that perform analysis workflow actions.
Non-executable comments are like a “note to self” that tells researchers what the code is doing and why. These annotations remind the researcher who originally wrote the code of the analytical workflow they used to generate their research results, while also making this information clear to other researchers. Code comments that support reproducibility often include explanations of what lines or blocks of code are meant to do or what outputs the code should produce.
The syntax for non-executable comments often uses a specified symbol or combination of symbols (*, //, /**, or # depending on the programming language) placed before the comment text, which signals to the software program that the text should be ignored.
The example below show examples of non-executable comments inserted in the code:
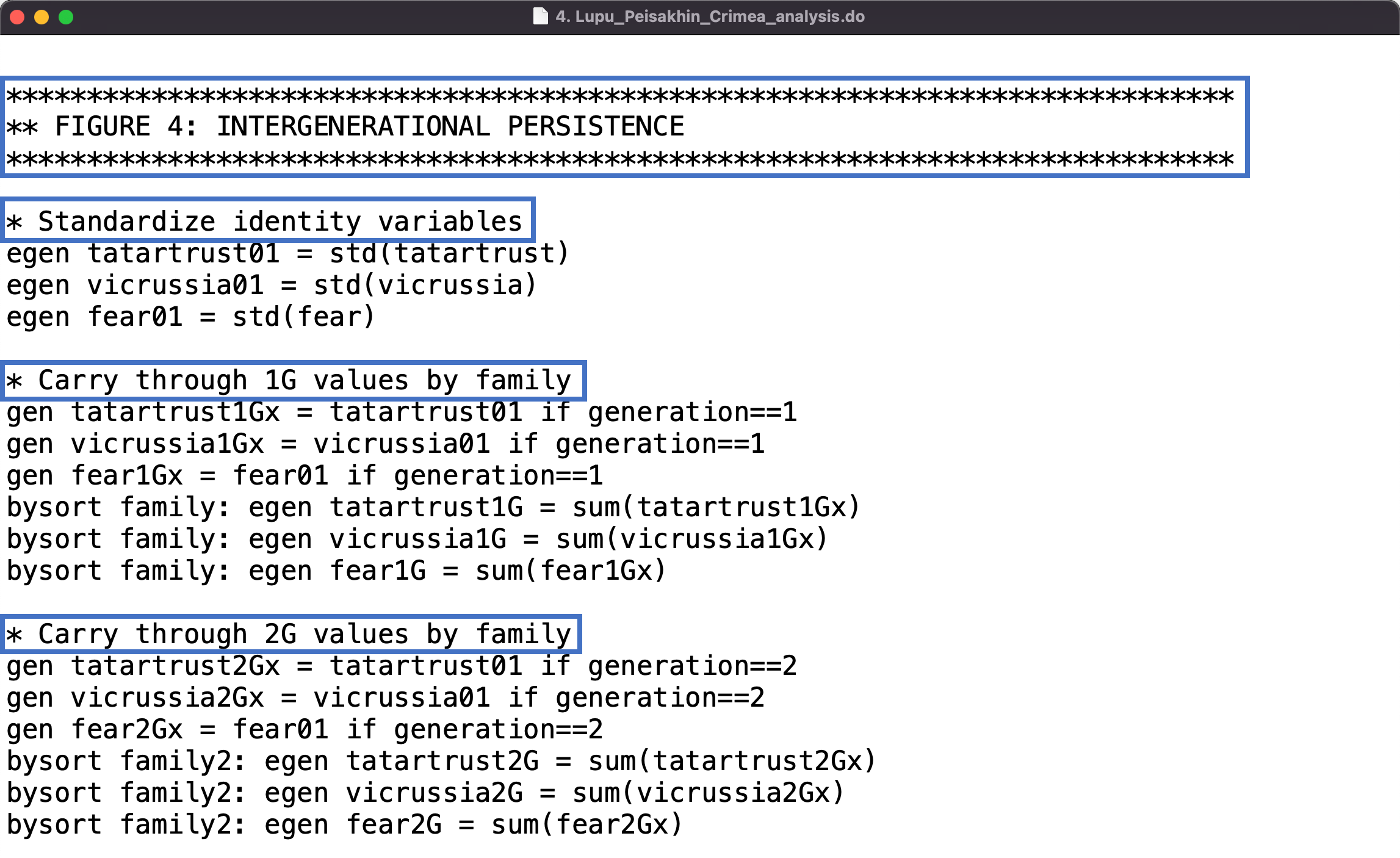
Spotlight: Sometimes Less is More
During the research process, it is not uncommon for blocks of code used to process and analyze data to gradually grow in size as researchers rework or broaden their approach. Oftentimes, these code blocks end up including lines of code that are no longer required to run the analysis and possibly produce unnecessary outputs.
Some researchers may decide that the unneeded code has some potential to be useful, whether to document previously used analytical steps or to be used in subsequent analyses. In these cases, these lines of code are not deleted, but instead commented out so that they are no longer executable.
On the other hand, if the code has no role in the analysis and no potential value, it is preferable that this extraneous code be deleted entirely to reduce the amount of lines of code that need to be inspected.
The Main File
In some cases, the analysis workflow is complex enough to require more than one code file to produce results. For research compendia that contain more than one code file, it is highly recommended to use file names that indicate the order in which they must be run and/or provide this information in the README file (see Episode 3: Documentation Review in Lesson 2: Curating for Reproducibility Workflows in the Curating for Reproducibility curriculum for more on the README file).
Use of a main file, while not a strict requirement, can do even more to enhance reproducibility. A main file, when executed, runs all of the other code files in the research compendium in the proper sequence to generate all of the tables, figures, and in-text results presented in the manuscript. It also provides an overview of how the code files are interconnected, which makes it easier to understand and re-trace the analytical workflow. This makes the main file the starting point for fully automated push-button reproducibility, which is what curating for reproducibility hopes to achieve.
Below is an example of code contained in a main file that includes commands to run code files in the proper order:
sysdir set PLUS ../Prerequisites
*ssc install estout
*ssc install xtqreg
do "1_1_household level data set.do"
do "1_2_multiple respondents per household.do"
do "2_1_merging hh level data all countries.do"
do "2_2_merging multiple responses data all countries.do"
do "3_1_merging all data sets all countries.do"
do "4_Main analysis.do"
do "5_SI Appendix.do"
log close
Spotlight: Our Language Choices
The main file may be more familiar to some people as the master file. While master file is the term that has been adopted to refer to the file containing code that initiates execution of code files in the correct order, this term is falling out of favor.
Many in the tech community are recognizing the terms such as “master” and “slave” are racially-loaded terms that can be offensive to Black people, who are underrepresented in tech community. By adopting more neutral terms such as main, primary, or parent for master and secondary, follower or child for slave, unnecessary references to slavery is avoided.
Such attention to language choices is an important step towards increasing diversity and inclusion in tech and in other disciplinary domains.
Software Dependencies
In some cases, researchers have found that statistical software programs lack functions for performing certain analytical operations. Adept researchers have written custom code themselves and organized it into a reusable package for sharing. This allows other researchers using the same analytical approaches to access and install the package into their own computational environments.
If a study requires the use of certain packages to run the analysis, the research compendium should document this software dependency. In addition, the code should not assume that users already have the packages installed and loaded. Rather, the code should include installation scripts for required packages.
When inspecting the code, look for commands that install and load packages into the program environment such as install.packages("package_name") or library(package).
Data Import
A key function of the code is to point the software program to the data to be used in the analysis, whether that data are included in the research compendium, or are accessed from an external source. The code inspection examines the commands for reading the data file(s) to determine if the code clearly indicates which data files are used in the analysis and where they are located. The inspection also checks to see whether the commands are written in a way that enables others to re-run the code without the need to make modifications to make it work correctly.
File paths
A file path is used to specify the location of a file within a directory structure. Take a look at where the analysisdata.csv file is located within the directory structure below:
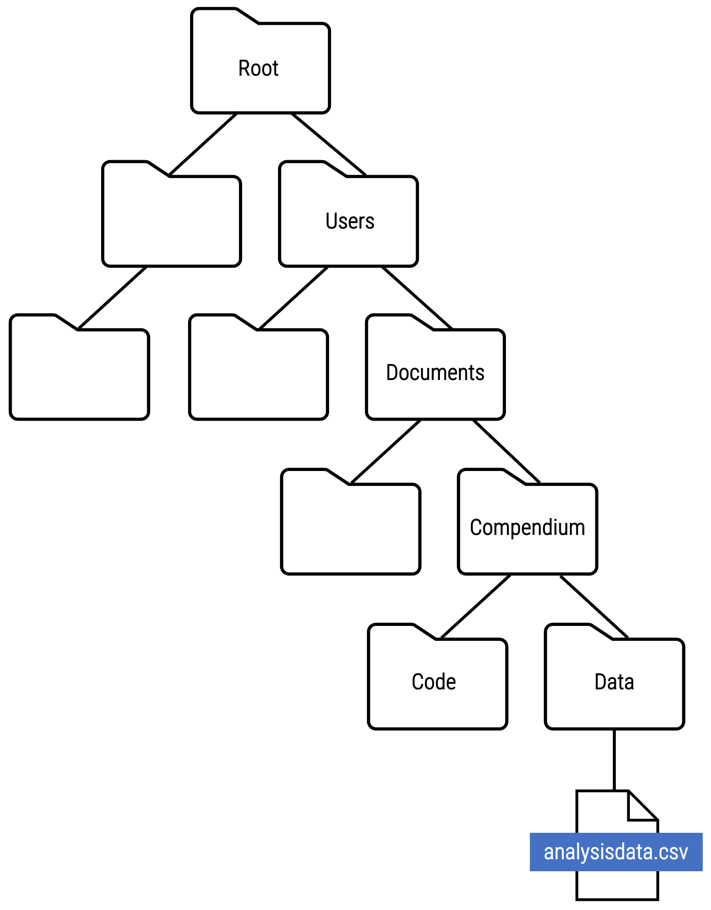
File paths are represented as a slash-separated list of the folder names followed by the file name, and can be written as either an absolute file path or relative file path.
Absolute file paths
C:\Users\Documents\Compendium\Data\ is an absolute file path that indicates the folder location of the analysisdata.csv file in relation to a specific root folder and all of the subfolders along the way.
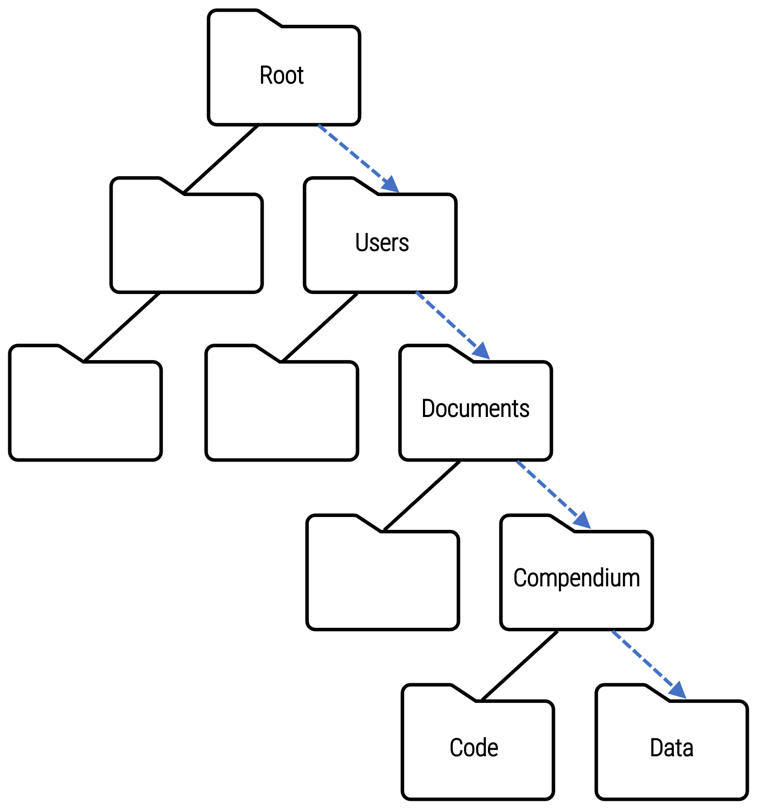
If the name or location of any one folder along the directory structure changes, the path will be broken, and the code will fail to read the data file. Moreover, using an absolute file path assumes that other users will have the same exact directory structure, which is not likely to be the case.
Relative file paths
../Compendium/Data indicates the folder location of the analysisdata.csv file relative to the current directory of the computing environment being used at the moment to run the code. This relative file path depends on folder names and locations of only part of the directory structure.
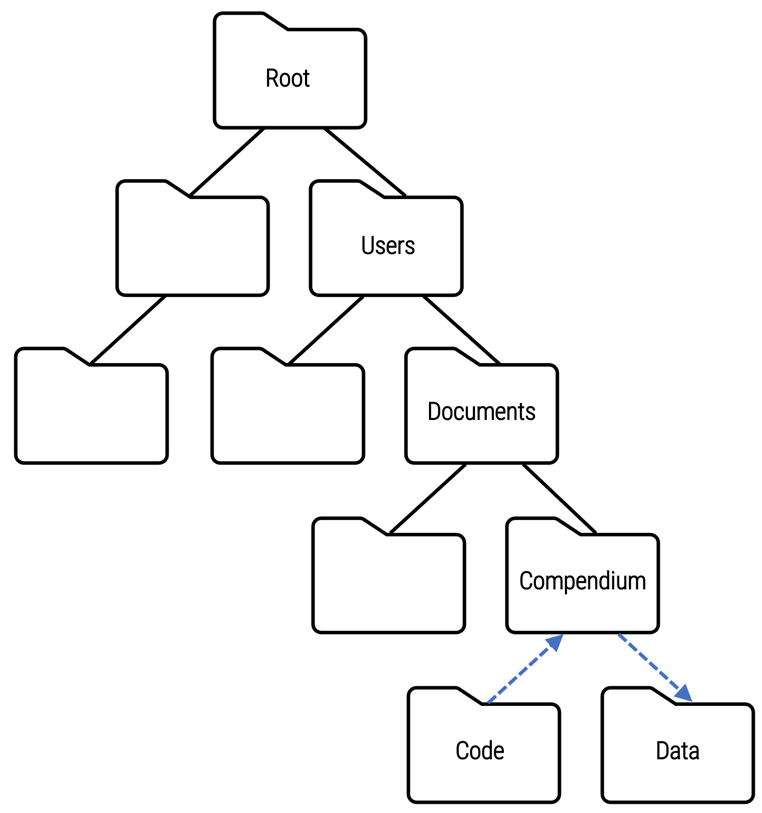
The recommended practice is to use relative file paths instead of absolute file paths because they are constructed in a way that facilitates reproducibility. Relative file paths anticipate re-use of the code by other researchers who will certainly use computing environments with varying directory structures.
Exercise: File Paths
To ensure that the analysis code executes successfully in any computing environment, relative file paths should be used to import data.
The R script to load data below uses an absolute file path. To support reproducibility, convert the absolute file path into a relative file path.
read.csv ("C:/Users/Documents/PopStudy1/Data/AnalysisData/pop1_analysis_data.csv")Solution
read.csv ("./PopStudy1/Data/AnalysisData/pop1_analysis_data.csv")
Putting It All Together
To assess whether the code upholds the reproducibility standard of being both independently understandable and reusable, code files must undergo an initial inspection to identify any issues that can make reproducibility difficult or impossible. The Code Inspection Checklist below outlines the specific tasks involved in a thorough inspection.
Checklist: Code Inspection
- Does the code file open and render properly?
- Does the code include the following components:
- Code header metadata
- Non-executable comments
- Package installation scripts
- Data import commands
- Variable transformation scripts
- Data analysis commands
- Log commands
- Does the code header metadata include all essential information needed to run the analysis (see the Code Header Metadata Checklist above)?
- Do non-executable comments provide signposts for the analytical steps in the computational workflow?
- Do data import commands use relative file paths instead of absolute file paths?
- If the research compendium contains multiple code files, do the filenames indicate the order in which the code files should be executed?
- If the research compendium contains multiple code files, is there a main file included in the compendium?
Key Points
Code inspection is the first step in reproducibility assessment. The inspection provides important information about the analytical workflow, computational requirements, and input data.
A main file contains essential code that facilitates reproducibility when more than one code file is required to repeat the analysis. The main file acts as the starting point in fully-automated push-button reproduction by running all code files in the proper sequence.
By provding the author’s contact information, computing requirements, and licensing terms, code header metadata signals to users of the author’s transparency and willingness to assist with reuse.
Non-executable comments in the code serve as a roadmap of the analytical workflow by identifying code segments and their purpose.
Code Execution
Overview
Teaching: 0 min
Exercises: 0 minQuestions
What does it mean to execute code in the context of a research compendium?
What can be done prior to publication to ensure that code will run error-free?
What are some of the common errors that cause non-executable code?
Objectives
Understand the importance of the code execution tasks to curating for reproducibility
Recall some of the common issues that prevent code from fully executing
Perform the code execution process as part of a reproduciblity assessment
After inspecting the code to be sure it includes all of the elements required to achieve reproducibility, the next step in the curating for reproducibility workflow is to run the code to confirm that it executes without errors and produces the outputs that match the results reported in the manuscript. This episode explains the process of running the code and describes some of the most common code errors encountered during reproducibility assessments.
Code Execution Overview
As the first reusers of the research compendium, curators can identify and address issues in the compendium that compromise standards for reproducibility before results are published in a report or manuscript. Not only does the curator inspect compendium files, but also, they verify that the files function as they should by using them to retrace the analytical workflow and reproduce reported results. This gives researchers assurances that research compendia they make available to the public meet high quality standards for reproducibility.
Executing the code provided in the research compendium is a critical step in assessing reproducibility, which requires that the research compendium be independently understandable and reusable. In practical terms, this means that anyone should be able to download the compendium and run the code to analyze the data and produce expected outputs–without having to modify the code or input data, and without assistance from the researcher(s) who originally wrote the code and performed the analysis.
Executing code as part of the code review process is not a common activity for many library and information professionals. However, adding this practice to curation and repository ingest workflows is imperative if reproducibility is the goal.
Running the Code
Prior to running the code, the computing environment will need to be set up according to the information provided in the research compendium. The initial code inspection, along with the data review, should give a good sense of the technical specifications for running the code.
When setting up the computing environment for running the code, consider the following:
Does the computing environment to be used to run the code meet the same or comparable technical requirements as described in the code header metadata (or in the README file)?
Differences in operating systems and software versions can yield discrepancies in outputs. Thus, the computing environment used to run the code for reproducibility assessment should match the provided specifications as closely as possible.
Does the code automatically install required packages or indicate in the header metadata how packages will need to be installed manually?
When running code to assess reproducibility, use the base (i.e., fresh) installation of the statistical software program without packages and libraries pre-installed. The base installation allows for detection of packages and libraries needed by the code to run correctly.
Can the code be run in a single sitting, or will the computation require more than one session?
Take note of the estimated run time required to execute the code. Complex or resource-heavy computations or analyses that use big data can take a significant time to run from beginning to end.
Spotlight: Capturing Information About the Computing Environment
There are specific commands that can help provide information about the computing environment or session while also identifying the packages necessary to run the code properly. While this information should be included in the code header metadata and/or elsewhere in research compendium documentation, curators can use these code commands to obtain this information if it is missing from the compendium.
# Print version information about R, the OS, and packages sessionInfo() # Print version information about the “pkg” package packageVersion(“pkg”)
Exercise: Let’s Run Some Code!
Open the sample code file and run the code. Did it run successfully?
Solution
If the code ran correctly using the specified software version, you should see this:
[IMAGE]
Spotlight: What About Containers?
Rather than enabling scripts to run on different machines, constraining the execution environment to a specific operating system and specific dependencies may make it easier to ensure re-execution.
A common approach to making it easier to rerun code is to do the computation on a cloud-based service or platform. Examples of this approach include WholeTale, Code Ocean, and MyBinder. Many of these are services are built on top of JupyterHub or RStudio, which encapsulate the compute environment in a container.
For desktop based workflows, the computation environment can be fixed in place using a number of solutions such as using a virtual machine or containerisation. This can be done by carefully constructing images or build scripts to use Vagrant, Docker or Singularity, usually starting with an image or container that includes all or most of the software required.
The image or container might be a suitable target for archiving (being mindful of licensing restrictions), or the scripts and/or config files that describe how to build the image or container, fetch the appropriate code and data, and then combine them is a potential approach for longer-term preservation of research compendia.
Common Code Issues
There are many reasons that code may not run properly. Oftentimes, it is not the fault of the researcher who originally wrote the code. As mentioned previously, differences in operating systems may affect the mechanics of the computation or cause discrepancies in the computational outputs. Software is often updated with bug fixes, new features, and other changes in ways that do not allow for backwards compatibility.
Other reasons that code may not execute fully (or not at all) may have something to do with the way the code was written. Below are some of the issues that are caused by faulty code.
Syntax errors
A simple typo in lines of code can cause syntax errors that cause code execution to fail. Running the code from beginning to end will catch these easy-to-fix errors.
Missing comments
Code that is written with reproducibility in mind will include non-executable comments that map code blocks to the tables, figures, and in-line results presented in the publication. The absence of such signposts, which will not necessarily cause code execution errors, still make reproducibility assessment cumbersome.
Use of absolute file paths
Absolute file paths assume that re-users have on their computer workstation a file directory structure identical to that of the original researcher. When it is not the case that the file directory is identical, running the code will result in an error indicating that the file cannot be found. Using relative paths makes the research compendium portable by calling files relative to its location in the current working directory.
# Read in the data file using an absolute file path
read.csv ("C:/Users/Documents/PopStudy1/Data/AnalysisData/pop1_analysis_data.csv")
# Read in the data file using an absolute relative file path.
read.csv ("./PopStudy1/Data/AnalysisData/pop1_analysis_data.csv")
Missing package installation scripts
Scripts to install packages are required to successfully execute the code (i.e., prerequisites). Without package installation scripts, the code will fail to execute until the packages are installed and loaded.
* Create Prerequisite folder and put cluster2.ado in this folder
sysdir set PLUS ..\Prerequisites
ssc install outreg2
ssc install wyoung
Missing seed
Any computation that generates random numbers (e.g., Monte Carlo simulations) requires a set seed to initialize the algorithm that generates the random numbers. Without that specific seed, the code will generate different random numbers, which will produce different outputs each time the code is run.
# Below, no seed is set so that every time the code is run, the output will be different.
# set year list
yearsets <- split(sample(years, length(years)), cut(seq(1,length(years)), breaks-ks, labels=FALSE))
# Below, the seed is set so that every time the code is run, the output will be the same.
# set year list
set.seed(123) # set seed for random number generation
yearsets <- split(sample(years, length(years)), cut(seq(1,length(years)), breaks-ks, labels=FALSE))
Exercise: Troubleshooting Problem Code
Run the code file that contains faulty code.
Answer the following questions:
- What issues did you encounter when executing the code?
- How might the issues be resolved so that the code runs successfully and produces the anticipated outputs?
Solution
solution
Spotlight: Coding Best Practices
Even code that executes properly can use some improvements to make it more readable and more efficient or elegant. This in turn can enhance reproducibility by making code that may appear to be “messy” to some users easier to understand and follow the sequence of analytical operations.
One way that code can be made more readable is to address how the code represents the sequence of operations so that the presentation of code blocks makes logical sense. Beyond inserting non-executable comments throughout the code to indicate what lines of code are meant to do or what the code is meant to produce, presenting code blocks in the same order in which corresponding results appear in the manuscript (if feasible to do so), makes it less cumbersome to assess reproducibility.
Another opportunity to improve code is to address inefficiencies in the code. A script may include commands that achieve a specific outcome, but do so inefficiently. For example, a script that includes repeated statements when other expressions are more appropriate like in the example below:
* Inefficient code SFA = SSMR1_N + SSPM1_N + SSST1N + SSAR1_N + SSDA1_N + SSLG1_N; if SSMR1_N = . then SFA = .; if SSPM1_N = . then SFA = .; if SSST1_N = . then SFA = .; if SSAR1_N = . then SFA = .; if SSDA1_N = . then SFA = .; if SSLG1_N = . then SFA = .; * Efficient code that achieves same output as inefficient code above SFA = sum(SSMR1_N, SSPM1_N, SSST1N, SSAR1_N, SSDA1_N, SSLG1_N) SFA = sum(SSMR1_N, SSPM1_N, SSST1N, SSAR1_N, SSDA1_N, SSLG1_N na.rm = TRUE)
Key Points
Executing code tests the reusability of the research compendium, which is a fundamental criterion of reproducibility.
Prior to running the code, the computing enviroment will need to meet the same or comparable techinical requirements as described in compendium documentation.
Simple errors that prevent code from fully executing can be easily addressed.
Output Review
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Why is output review an essential component of curating code for reproducibility?
What are outputs and why are they important?
What are the essential steps in output and manuscript review?
Objectives
Understand the importance of code output and manuscript review to curating for reproducibilty practice
Compare code outputs with manuscript results to identify discrepancies
Understand how to address inconsistencies found during code output and manuscript review
You reviewed files, documentation, and data. You ran the code and confirmed that it is error-free. You are now ready for a key part of curating for reproducibility: comparing the output produced by the code with the findings reported in the article!
Manuscript Results
An initial step in assessing reproducibility is inspecting the manuscript to identify all analysis results appearing in tables, graphs, figures, and in-text references. For the purposes of this lesson, we use the term “manuscript” to refer to a working paper, draft, preprint, article under review, or a published article.
Inspecting the manuscript requires a close reading of the entire document, which may include appendices and supplementary materials. The goal of this task is to confirm that the code includes the commands needed to reproduce tables and figures, as well as results appearing as in-line text and not referenced in tables, figures, and/or graphs.
To facilitate this process, curators may highlight sections of the manuscript where results appear. Doing so will make it easier to compare code outputs to the results in the manuscript. The next exercise offers an opportunity to practice this technique.
Exercise: Identifying Manuscript Results
Perform a manuscript inspection by doing a close reading of the entire manuscript and highlighting sections of the manuscript that present analysis results. Be sure that you highlight all figures, tables, graphs, and in-text numbers.
Solution
solution
Code Outputs
When code executes successfully, the program will display the results of the computations, i.e., code outputs. Sometimes, these outputs can be difficult to interpret by non-domain experts, or simply because the outputs lack the neat formatting of tables, figures, and graphs seen in the manuscript. In other cases, code may generate outputs in an order that does not align with the orientation of results in the manuscript. Output files and log files can make it easier to locate and interpret code outputs for code output and manuscript review purposes.
Output files
Depending on how it is written, the code may include commands or scripts that produce output files that contain an image of a graph, figure, table, or some other result, which is often embedded in the manuscript. These output files are standalone artifacts that are included in the research compendium. Below are some output file examples:
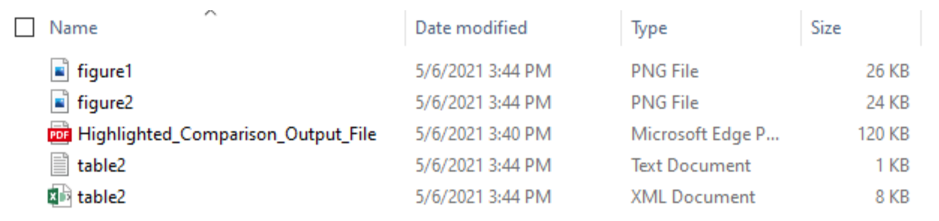
The log file
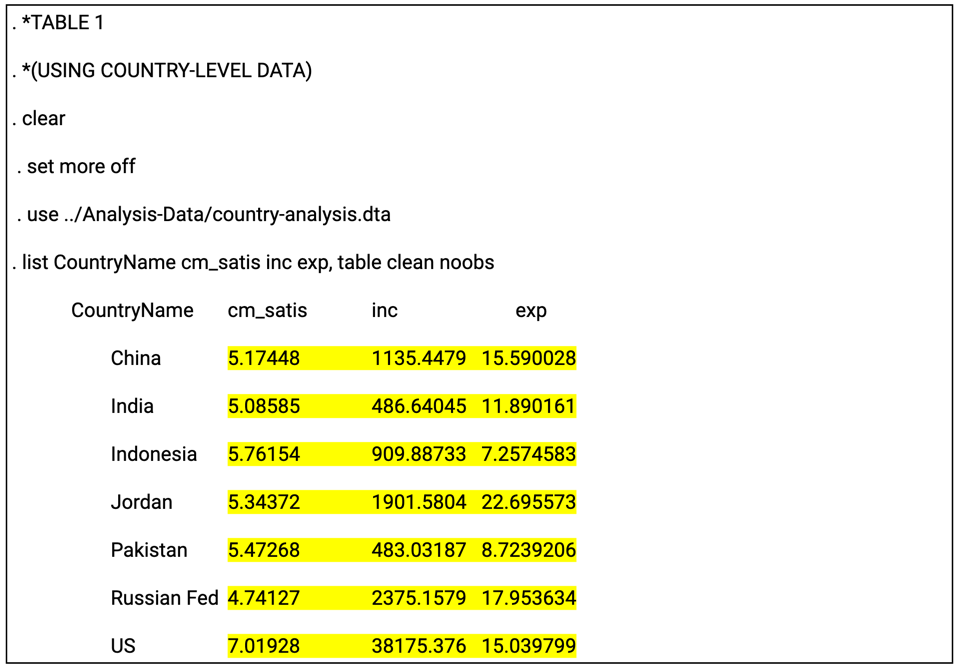
The log file is of particular importance in the reproducibility assessment process because it not only can present code outputs in a readable format, but also it serves as a record of the analytical workflow. The log file provides a written log of the computational events that occurred during a given session during which the program was executed. The log file can be generated automatically or manually, and typically serves as a reference for researchers as they write or revise the manuscript. Below is an example log:
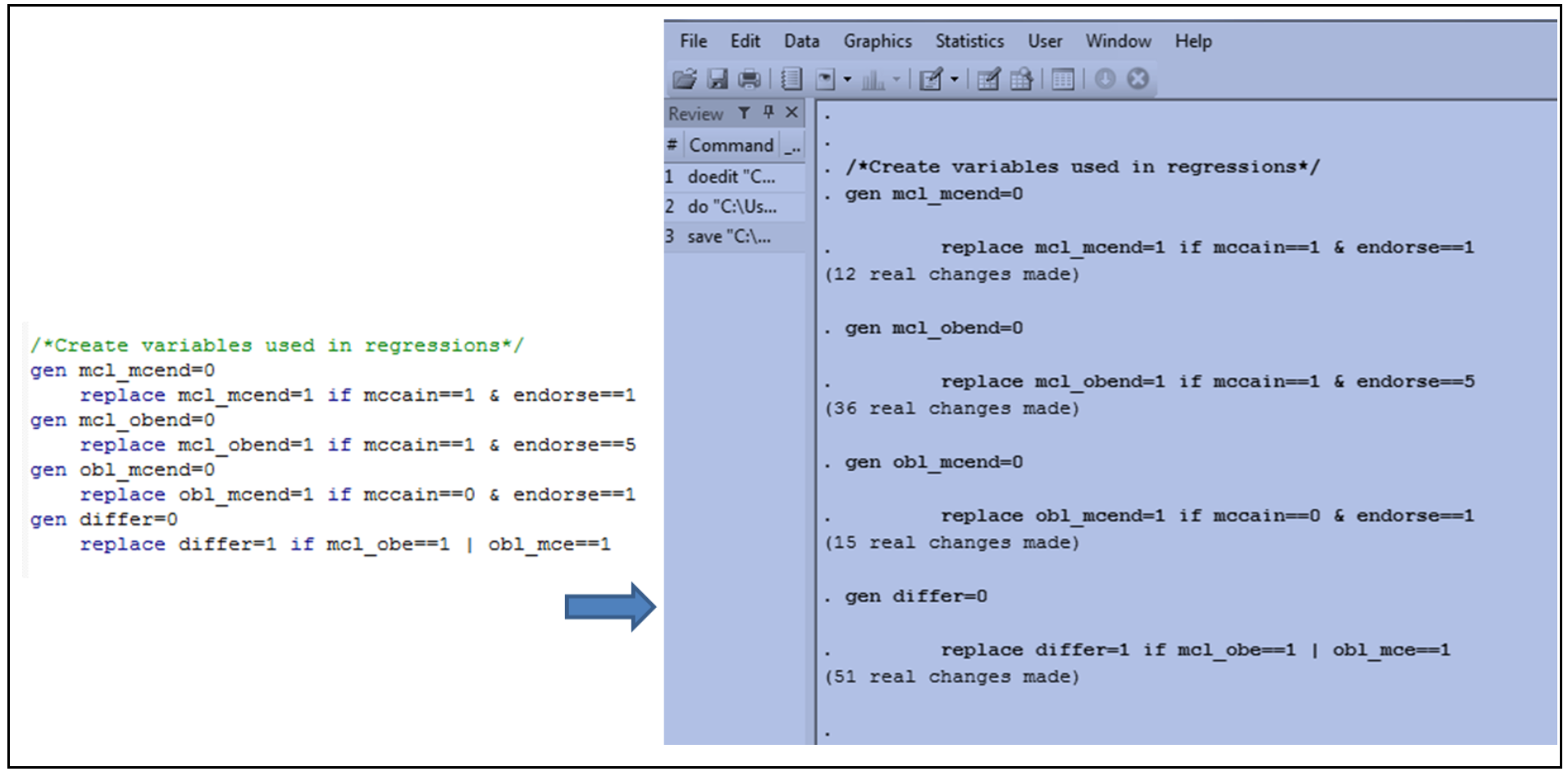
For curators, the log file can be useful because results are written into the log (with the exception of graphs), which can be used to reference code outputs when assessing whether or not the research compendium can be used to reproduce manuscript results. The image below shows an example of code and a section of the log file that the code produced:
Spotlight: When Data are Restricted
For some research compendium, data files cannot be included because they contain personally identifiable informationn (PII), protected health information (PHI), or otherwise restricted data that cannot be shared publicly, even for curation purposes. In these cases, the log file can provide evidence that the computational workflow was executed, and that it produced the outputs featured in the manuscript.
Like much of what we have been learning, descriptions of concepts and processes can seem abstract when reading about them alone. Making the connection from concept to practice takes time, and cultivating a new skill takes practice. Let’s practice what we have learned in Lesson 3 thus far by digging into outputs from a real life study.
Identifying Code Outputs
In the sample research compendium, the researcher has included a log file that includes code outputs. Review the log closely to identify code outputs. Highlight all sections of the log that show code outputs.
Solution
solution
Comparing Code Outputs to Reported Results
The examples below show the output of specific code blocks alongside their corresponding results presented in the manuscript.
Crosstab and Chi-Square (Stata):
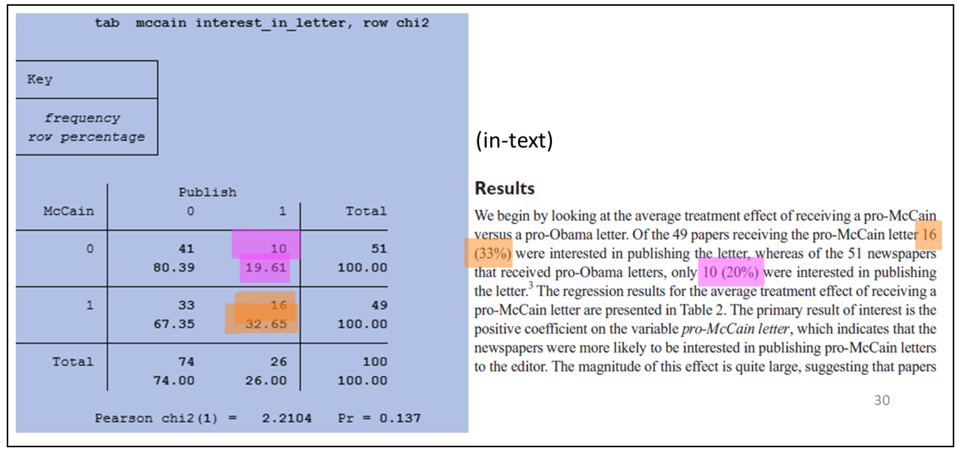
Probit regression (Stata):
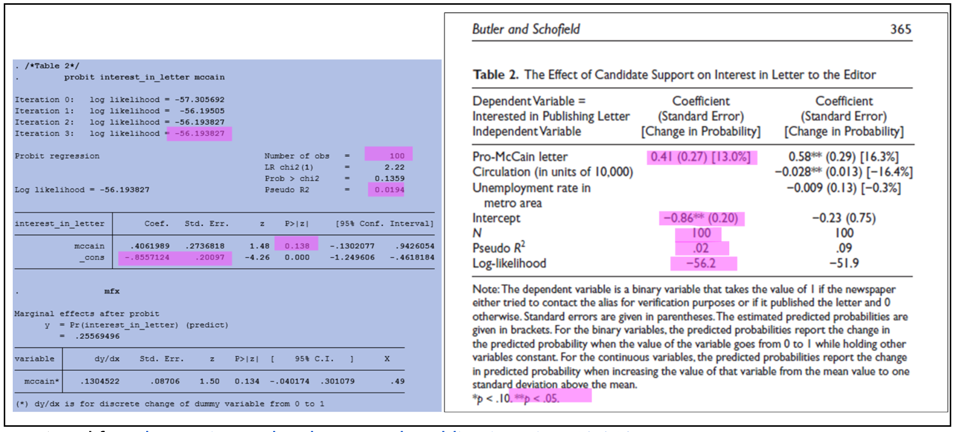
Verifying Reproducibility
Verifying reproducibility requires that the results reported in the manuscript are compared against outputs generated by the code. Any inconsistencies found when comparing code outputs to corresponding results in the manuscript indicates irreproducibility.
Inconsistencies can appear in several ways, from differences in decimal rounding and mislabeled graphs, to more significant discrepancies as shown in the example below:
Exercise: Comparing Code Outputs to Manuscript Results
Using the manuscript and log file from the previous exercises, compare the code outputs highlighted in the log file and the results highlighted in the manuscript. List any discrepancies you find between code outputs and corresponding results in the manuscript.
Solution
solution
The checklist below outlines the tasks completed during the code output and manuscript review as part of the code review component of the curation for reproducibility workflow.
Checklist: Code Output and Manuscript Review
- Review the manuscript, including appendices and supplemental materials, to locate and highlight analysis results displayed in figures, tables, graphs, and in-text numbers.
- Review the log file to locate and highlight code outputs.
- Match the highlighted code outputs in the log file to the highlighted results in the manuscript.
- Compare code outputs to manuscript results to confirm an exact match of numerical results
- If output files are included in the compendium, compare figures, graphs, and tables in the output files to the figures, graphs, and tables in the manuscript.
- Document any discrepancy–no matter how minor–found between the code outputs and manuscript results
Addressing Inconsistencies
When inconsistencies are discovered during the code output and manuscript results comparison, they should be documented in enough detail so that the researcher is able to locate the discrepancy and take corrective actions to resolve the inconsistencies.
It is highly recommended that researchers run their code and verify their results themselves prior to submitting their compendium for curator review. Researchers know their research best and can spot and address problems more quickly than any third party can.
More often than not, there are simple explanations for these inconsistencies. The vast majority of researchers share their compendia in good faith and with the expectation that their code will reproduce their reported results. As the first re-users of the research compendium, curators can flag these inconsistencies so that researchers have the opportunity to make corrections before sharing it publicly.
Continue to Lesson 4: Compendium Packaging and Publishing of the Curating for Reproducibility curriculum to learn about considerations for sharing the research compendium.
Key Points
Every table, figure, and in-text number in the manuscript should be accounted for in code outputs.
The curating for reproducibilty workflow includes identifying specific commands in code files that produce outputs, and then verifying that reported results can be reproduced using that code.
Curators should document discrepanices found during the code output and manuscript review process and take steps to address them.