Compendium Packaging
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Why is it important to package research artifacts into a compendium?
What are best practices for packaging files into the research compendium?
How does the TIER Protocol help with compendium packaging?
Objectives
Explain the importance of organizing files in the research compendium
Apply the TIER Protocol to organize files within a research compendium
Research Compendium Packaging
When preparing research materials to submit to a trustworthy repository for long-term preservation and sharing, it is important to package the materials in a way that enables other researchers to easily understand the contents of each artifact contained in the compendium and their function in relationship to one another. This can be accomplished with logical file organization, standard file naming schemes, and file version control.
File Organization: The TIER Protocol
The TIER (Teaching Integrity in Empirical Research) Protocol offers a framework that specifies which files should be included in a research compendium, what those files should contain, and how those files should be organized in a hierarchical file folder structure. The TIER Protocol was designed to support four specific standards for computational reproducibility:
-
Sufficiency. The research compendium should contain all materials necessary to computationally reproduce reported results.
-
Soup-to-nuts. The research compendium should contain all materials necessary to re-trace the computational workflow from beginning (preparing data for analysis) to end
-
Portability. The research compendium should continental all materials necessary to allow for computational reproduction on any computer workstation.
-
(Almost) one-click reproducibility. The research compendium should contain all materials necessary to generate results using a single master script.
As discussed in previous lessons in the Curating for Reproducibility Curriculum, the research compendium, at a minimum, should contain a readme, dataset, codebook, and script file. The file folder granularity of TIER Protocol 4.0 goes further to reflect the reality of typical computational research that often requires several data and script files to perform all of the steps in the analytical workflow, some of which may generate intermediate files that then become part of the analysis to produce outputs.
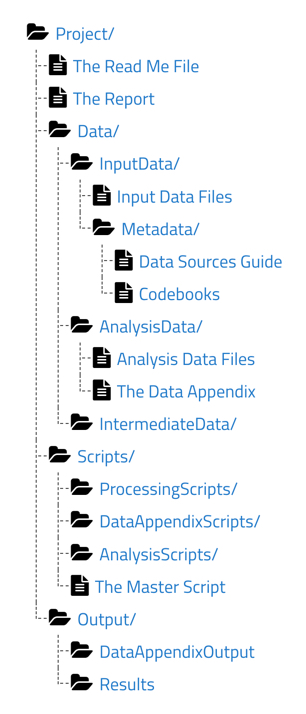
Within the root Project/ folder are the README file (see Episode 3: Documentation Review] in Lesson 2: Curating for Reproducibilty Workflows of the CuRe Curriculum), the Report file (final article/report), and three primary subfolders each containing specific files as described below.
Data/
This folder should contain input data files containing the data obtained from a primary source or constructed from data collection/generation efforts, analysis data files containing the processed data used in the analysis, and any intermediate data files created during data processing and stored temporarily to be used in subsequent steps of the analysis.
In addition, for each input data file, there should be included a codebook that defines the variables in the dataset and a data sources guide that provides a data citation, data availability/licensing information, and instructions on how to access the data from their original sources.
For each analysis data file, the TIER Protocol also requires a data appendix. Similar to a codebook, the data appendix defines each variable. However, the data appendix also provides information on the unit of observation, basic summary statistics, and variable distributions.
Scripts/
This folder should include processing scripts used to transform input data into analysis data files, analysis scripts used to generate results presented in the final report/article, and data appendix scripts used to produce results presented in the appendix of the final report/article.
To help achieve (almost) one-click reproducibility, the TIER Protocol also recommends a master script file, or main script file, (see Episode 3: Code Inspection in Lesson 3: Reproducibility Assessment of the CuRe Curriculum) that runs all of the scripts in the proper order with as little human intervention as possible beyond executing the main script to produce the results presented in the report/article and the appendix.
Output/
This folder should contain results files that include the figures, tables, and any other results presented in the final report/article as well as data appendix output files that include the figures tables and any other outputs presented in the appendix of the final report/article.
Learn more about the TIER Protocol here:
https://www.projecttier.org/
It is the rare researcher who routinely keeps their compendium files in a neat folder structure like the one suggested by the TIER Protocol. More often, files are delivered in a single folder with little information on what each file contains. The next challenge will give you a chance to transform a collection of files into an organized research compendium.
Exercise: TIER(s) of Joy
As this episode explained, organizing files in a logical manner supports reproducibility by making it more evident the purpose of each file and the relationships among them. This is useful during curation for reproducibility workflows that require understanding of the analytical workflow in order to re-execute the analysis to confirm reproducibility of results.
The file list below was extracted from the original README.txt included in a research compendium submitted for curation. Organize the study files in a logical folder structure as outlined by TIER Protocol 4.0.
Solution
solution
File Naming
A good filename gives anyone who encounters the file an immediate sense of what it contains and its function among other related files. By establishing and consistently applying a standard file naming convention, researchers, collaborators, and future users of the research compendium can more easily locate, understand, and use the compendium files.
Filename elements
An effective file naming convention prescribes standardized filenames composed of descriptive elements that uniquely and concisely identify each file. A convention might require some of the following elements:
- short study title or study acronym
- study location
- sample
- content type
- file production/update date
- researcher initials
- file version number
Filename formatting
File naming conventions should also take into account the computing environment in which the files will be rendered and used. Some operating systems and software applications impose character limits for object names or treat characters in peculiar ways during certain operations. To avoid problems as a result of this, file naming conventions should also establish formatting standards for filenames and specific elements within filenames such as those below:
- Keep filenames as short as possible.
- Avoid the use of spaces and special characters.
- Use underscores or camel case to enhance human readability.
- Apply the ISO 8601 (YYYYMMDD) format for dates.
- Include leading zeros for sequential file numbering.
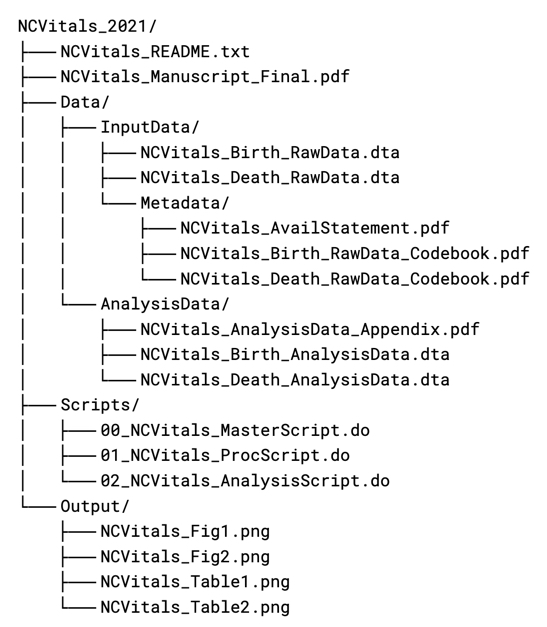
The example below shows the application of a file naming convention to files contained in a research compendium.
Version Control
During the course of the research lifecycle, data are being manipulated, scripts are being updated, and documentation is being revised. Recurrent changes made to files in the research compendium reflect the iterative nature of research, which results in the accumulation of several versions of a given file. Version control not only keeps track of each version to make it easy to identify the precise active files that should be used to perform a task, but also it makes it possible to revert to prior versions of a file should issues with the active file make it necessary. With regard to computational reproducibility, being able to identify the precise files to be used to execute the computational workflow to generate reported results is critical.
Version control can be as simple as appending standardized file version information to the file name, with the final version limited to read-only access.
NCVitals_Birth_AnalysisData_20210213_v01.dta
NCVitals_Birth_AnalysisData_20210216_v02.dta
NCVitals_Birth_AnalysisData_20210522_v03.dta
NCVitals_Birth_AnalysisData_20210629_v04FINAL.dta
For research processes that require frequent file updates, the use of a file storage and management system that automates the assignment of version numbers to files as they are created and updated (e.g., Box, Open Science Framework) can reduce the effort of tracking versions of files.
The often non-linear nature of computational research can make version control of script files a difficult task. Version control systems like Git, GitHub, and Bitbucket are becoming more popular among researchers as a tool for tracking changes to script files as they write and revise their code. These tools have automated processes for storing iterations of files while also capturing information about each iteration (when it was created, who created it, what changes were made to it). Git is especially useful for collaborative projects because it also helps to synchronize programming activities among collaborators, who otherwise would require careful coordination to ensure the correct files are being used.
It is important to put into practice what one learns as they learn it to not only retain but begin making it a habit. Let’s try it here by practicing file naming.
Exercise: What’s in a (File) Name?
How would you rename the files below to make it easier for researchers to have a better sense of their contents and function, be able to track file versions, and ensure the filenames are machine-readable?
parent-add health study wave 3_nov19.csvaddhealth_child_w3_10-24-22.csvchild-add health study-wave3 final.csvSolution
addhealth_w3_parent_20221119.csvaddhealth_w3_child_20221024.csvaddhealth_w3_child_final.csv
Key Points
It is important to package research artifacts into a research compendium that uses a logical file folder structure and standard file naming conventions to enable others to make sense of the files and the functional relationships among them.
The TIER Protocol is a useful framework for organizing compendium files that supports four essential computational reproducibility standards: sufficiency, soup-to-nuts, portability, and (almost) one-click reproducibility.
Research is a non-linear process that often requires frequent file revisions. Version control is critical to ensure that the files included in the compendium are the precise ones that can be used to reproduce reported results.